Probabilistic Forecasting: Conformal Prediction¶
Conformal prediction is a framework for constructing prediction intervals that are guaranteed to contain the true value with a specified probability (coverage probability). It works by combining the predictions of a point-forecasting model with its past residuals—differences between previous predictions and actual values. These residuals help estimate the uncertainty in the forecast and determine the width of the prediction interval that is then added to the point forecast. Skforecast implements Split Conformal Prediction (SCP).
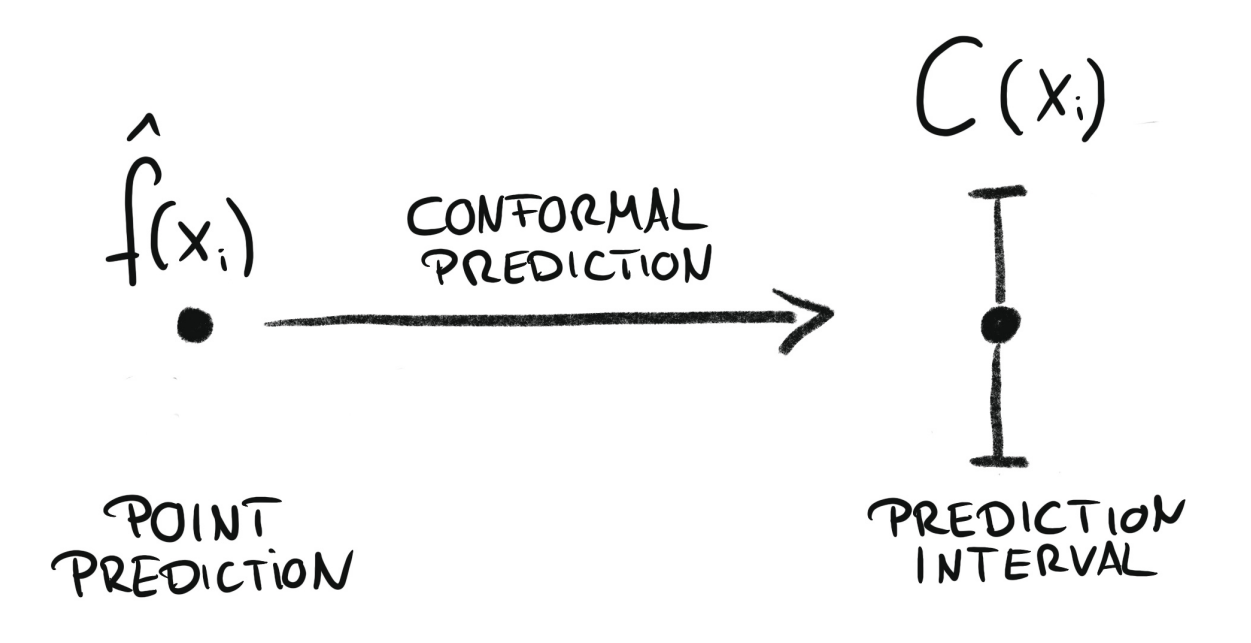
Conformal regression turns point predictions into prediction intervals. Source: Introduction To Conformal Prediction With Python: A Short Guide For Quantifying Uncertainty Of Machine Learning Models
by Christoph Molnar https://leanpub.com/conformal-prediction
Conformal methods can also calibrate prediction intervals generated by other techniques, such as quantile regression or bootstrapped residuals. In this case, the conformal method adjusts the prediction intervals to ensure that they remain valid with respect to the coverage probability. Skforecast provides this functionality through the ConformalIntervalCalibrator transformer.
⚠ Warning
There are several well-established methods for conformal prediction, each with its own characteristics and assumptions. However, when applied to time series forecasting, their coverage guarantees are only valid for one-step-ahead predictions. For multi-step-ahead predictions, the coverage probability is not guaranteed. Skforecast implements Split Conformal Prediction (SCP) due to its simplicity and efficiency.
💡 Tip
For more examples on how to use probabilistic forecasting, check out the following articles:
Libraries and data¶
# Data processing
# ==============================================================================
import numpy as np
import pandas as pd
from skforecast.datasets import fetch_dataset
# Plots
# ==============================================================================
import matplotlib.pyplot as plt
from skforecast.plot import set_dark_theme, plot_residuals, plot_prediction_intervals
# Modelling and Forecasting
# ==============================================================================
from lightgbm import LGBMRegressor
from skforecast.recursive import ForecasterRecursive
from skforecast.preprocessing import RollingFeatures
from skforecast.model_selection import TimeSeriesFold, backtesting_forecaster
from skforecast.metrics import calculate_coverage
# Configuration
# ==============================================================================
import warnings
warnings.filterwarnings('once')
# Data
# ==============================================================================
data = fetch_dataset(name="ett_m2_extended")
data = data[[
"OT",
"day_of_week_cos",
"day_of_week_sin",
"hour_cos",
"hour_sin",
"month_cos",
"month_sin",
"week_cos",
"week_sin",
"year",
]]
data.head(2)
ett_m2_extended --------------- Data from an electricity transformer station was collected between July 2016 and July 2018 (2 years x 365 days x 24 hours x 4 intervals per hour = 70,080 data points). Each data point consists of 8 features, including the date of the point, the predictive value "Oil Temperature (OT)", and 6 different types of external power load features: High UseFul Load (HUFL), High UseLess Load (HULL), Middle UseFul Load (MUFL), Middle UseLess Load (MULL), Low UseFul Load (LUFL), Low UseLess Load (LULL). Additional variables are created based on calendar information (year, month, week, day of the week, and hour). These variables have been encoded using the cyclical encoding technique (sin and cos transformations) to preserve the cyclical nature of the data. Zhou, Haoyi & Zhang, Shanghang & Peng, Jieqi & Zhang, Shuai & Li, Jianxin & Xiong, Hui & Zhang, Wancai. (2020). Informer: Beyond Efficient Transformer for Long Sequence Time-Series Forecasting. [10.48550/arXiv.2012.07436](https://arxiv.org/abs/2012.07436). https://github.com/zhouhaoyi/ETDataset Shape of the dataset: (69680, 16)
| OT | day_of_week_cos | day_of_week_sin | hour_cos | hour_sin | month_cos | month_sin | week_cos | week_sin | year | |
|---|---|---|---|---|---|---|---|---|---|---|
| date | ||||||||||
| 2016-07-01 00:00:00 | 38.661999 | -0.900969 | -0.433884 | 1.0 | 0.0 | -0.866025 | -0.5 | -1.0 | 1.224647e-16 | 2016 |
| 2016-07-01 00:15:00 | 38.223000 | -0.900969 | -0.433884 | 1.0 | 0.0 | -0.866025 | -0.5 | -1.0 | 1.224647e-16 | 2016 |
# Split train-calibration-test
# ==============================================================================
exog_features = data.columns.difference(['OT']).tolist()
end_train = '2017-10-01 23:59:00'
end_calibration = '2018-04-01 23:59:00'
data_train = data.loc[: end_train, :]
data_cal = data.loc[end_train:end_calibration, :]
data_test = data.loc[end_calibration:, :]
print(f"Dates train : {data_train.index.min()} --- {data_train.index.max()} (n={len(data_train)})")
print(f"Dates calibration: {data_cal.index.min()} --- {data_cal.index.max()} (n={len(data_cal)})")
print(f"Dates test : {data_test.index.min()} --- {data_test.index.max()} (n={len(data_test)})")
Dates train : 2016-07-01 00:00:00 --- 2017-10-01 23:45:00 (n=43968) Dates calibration: 2017-10-02 00:00:00 --- 2018-04-01 23:45:00 (n=17472) Dates test : 2018-04-02 00:00:00 --- 2018-06-26 19:45:00 (n=8240)
# Plot partitions
# ==============================================================================
set_dark_theme()
plt.rcParams['lines.linewidth'] = 0.5
fig, ax = plt.subplots(figsize=(8, 3))
ax.plot(data_train['OT'], label='Train')
ax.plot(data_cal['OT'], label='Calibration')
ax.plot(data_test['OT'], label='Test')
ax.set_title('Oil Temperature')
ax.legend();
Calibration residuals¶
To address the issue of overoptimistic intervals, it is recomended to use out-sample residuals. These are residuals from a calibration set, which contains data not seen during training. These residuals can be obtained through backtesting.
# Create and fit forecaster
# ==============================================================================
params = {
"max_depth": 4,
"verbose": -1,
"random_state": 15926
}
lags = [1, 2, 3, 23, 24, 25, 47, 48, 49, 71, 72, 73]
window_features = RollingFeatures(stats=["mean"], window_sizes=24 * 3)
forecaster = ForecasterRecursive(
regressor = LGBMRegressor(**params),
lags = lags,
window_features = window_features,
)
forecaster.fit(
y = data.loc[:end_calibration, 'OT'],
exog = data.loc[:end_calibration, exog_features]
)
forecaster
ForecasterRecursive
General Information
- Regressor: LGBMRegressor
- Lags: [ 1 2 3 23 24 25 47 48 49 71 72 73]
- Window features: ['roll_mean_72']
- Window size: 73
- Exogenous included: True
- Weight function included: False
- Differentiation order: None
- Creation date: 2025-03-07 13:07:46
- Last fit date: 2025-03-07 13:07:47
- Skforecast version: 0.15.0
- Python version: 3.12.9
- Forecaster id: None
Exogenous Variables
-
day_of_week_cos, day_of_week_sin, hour_cos, hour_sin, month_cos, month_sin, week_cos, week_sin, year
Data Transformations
- Transformer for y: None
- Transformer for exog: None
Training Information
- Training range: [Timestamp('2016-07-01 00:00:00'), Timestamp('2018-04-01 23:45:00')]
- Training index type: DatetimeIndex
- Training index frequency: 15min
Regressor Parameters
-
{'boosting_type': 'gbdt', 'class_weight': None, 'colsample_bytree': 1.0, 'importance_type': 'split', 'learning_rate': 0.1, 'max_depth': 4, 'min_child_samples': 20, 'min_child_weight': 0.001, 'min_split_gain': 0.0, 'n_estimators': 100, 'n_jobs': None, 'num_leaves': 31, 'objective': None, 'random_state': 15926, 'reg_alpha': 0.0, 'reg_lambda': 0.0, 'subsample': 1.0, 'subsample_for_bin': 200000, 'subsample_freq': 0, 'verbose': -1}
Fit Kwargs
-
{}
# Backtesting on calibration data to obtain out-sample residuals
# ==============================================================================
cv = TimeSeriesFold(
steps = 24,
initial_train_size = len(data.loc[:end_train]),
refit = False
)
_, predictions_cal = backtesting_forecaster(
forecaster = forecaster,
y = data.loc[:end_calibration, 'OT'],
exog = data.loc[:end_calibration, exog_features],
cv = cv,
metric = 'mean_absolute_error',
n_jobs = 'auto',
verbose = False,
show_progress = True
)
0%| | 0/728 [00:00<?, ?it/s]
# Distribution of out-sample residuals
# ==============================================================================
residuals = data.loc[predictions_cal.index, 'OT'] - predictions_cal['pred']
print(pd.Series(np.where(residuals < 0, 'negative', 'positive')).value_counts())
plt.rcParams.update({'font.size': 8})
_ = plot_residuals(residuals=residuals, figsize=(7, 4))
negative 12251 positive 5221 Name: count, dtype: int64
With the set_out_sample_residuals() method, the out-sample residuals are stored in the forecaster object so that they can be used to calibrate the prediction intervals.
# Store out-sample residuals in the forecaster
# ==============================================================================
forecaster.set_out_sample_residuals(
y_true = data.loc[predictions_cal.index, 'OT'],
y_pred = predictions_cal['pred']
)
Now that the new residuals have been added to the forecaster, the prediction intervals can be calculated using use_in_sample_residuals = False.
# Prediction intervals
# ==============================================================================
forecaster.predict_interval(
steps = 24,
exog = data.loc[end_calibration:, exog_features],
interval = [10, 90],
method = 'conformal',
use_in_sample_residuals = False,
use_binned_residuals = True
)
| pred | lower_bound | upper_bound | |
|---|---|---|---|
| 2018-04-02 00:00:00 | 32.024773 | 29.387492 | 34.662053 |
| 2018-04-02 00:15:00 | 31.853564 | 29.216283 | 34.490845 |
| 2018-04-02 00:30:00 | 31.695225 | 29.057944 | 34.332506 |
| 2018-04-02 00:45:00 | 31.442913 | 28.805632 | 34.080194 |
| 2018-04-02 01:00:00 | 31.390724 | 28.753443 | 34.028005 |
| 2018-04-02 01:15:00 | 31.137807 | 28.500526 | 33.775088 |
| 2018-04-02 01:30:00 | 30.926586 | 28.289305 | 33.563867 |
| 2018-04-02 01:45:00 | 30.793334 | 28.156053 | 33.430615 |
| 2018-04-02 02:00:00 | 30.544763 | 27.907482 | 33.182044 |
| 2018-04-02 02:15:00 | 30.544763 | 27.907482 | 33.182044 |
| 2018-04-02 02:30:00 | 30.569183 | 27.931902 | 33.206464 |
| 2018-04-02 02:45:00 | 30.585077 | 27.947796 | 33.222358 |
| 2018-04-02 03:00:00 | 30.589153 | 27.951873 | 33.226434 |
| 2018-04-02 03:15:00 | 30.589153 | 27.951873 | 33.226434 |
| 2018-04-02 03:30:00 | 30.594971 | 27.957691 | 33.232252 |
| 2018-04-02 03:45:00 | 30.613403 | 27.976122 | 33.250683 |
| 2018-04-02 04:00:00 | 30.613403 | 27.976122 | 33.250683 |
| 2018-04-02 04:15:00 | 30.609986 | 27.972705 | 33.247267 |
| 2018-04-02 04:30:00 | 30.625439 | 27.988158 | 33.262719 |
| 2018-04-02 04:45:00 | 30.625439 | 27.988158 | 33.262719 |
| 2018-04-02 05:00:00 | 30.637124 | 27.999843 | 33.274405 |
| 2018-04-02 05:15:00 | 30.636440 | 27.999159 | 33.273721 |
| 2018-04-02 05:30:00 | 30.647419 | 28.010138 | 33.284699 |
| 2018-04-02 05:45:00 | 30.647419 | 28.010138 | 33.284699 |
It is also possible to use forecast prediction intervals within a backtesting loop.
# Backtesting with prediction intervals in test data using out-sample residuals
# ==============================================================================
cv = TimeSeriesFold(
steps = 24,
initial_train_size = len(data.loc[:end_calibration]),
refit = False
)
metric, predictions = backtesting_forecaster(
forecaster = forecaster,
y = data['OT'],
exog = data[exog_features],
cv = cv,
metric = 'mean_absolute_error',
interval = [10, 90], # 80% prediction interval
interval_method = 'conformal',
use_in_sample_residuals = False, # Use out-sample residuals
use_binned_residuals = True # Adaptive conformal
)
predictions.head(5)
0%| | 0/344 [00:00<?, ?it/s]
| pred | lower_bound | upper_bound | |
|---|---|---|---|
| 2018-04-02 00:00:00 | 32.024773 | 29.387492 | 34.662053 |
| 2018-04-02 00:15:00 | 31.853564 | 29.216283 | 34.490845 |
| 2018-04-02 00:30:00 | 31.695225 | 29.057944 | 34.332506 |
| 2018-04-02 00:45:00 | 31.442913 | 28.805632 | 34.080194 |
| 2018-04-02 01:00:00 | 31.390724 | 28.753443 | 34.028005 |
✎ Note
Two arguments control the use of residuals in predict() and backtesting_forecaster():
use_in_sample_residuals: IfTrue, the in-sample residuals are used to compute the prediction intervals. Since these residuals are obtained from the training set, they are always available, but usually lead to overoptimistic intervals. IfFalse, the out-sample residuals (calibration) are used to calculate the prediction intervals. These residuals are obtained from the validation/calibration set and are only available if theset_out_sample_residuals()method has been called. It is recommended to use out-sample residuals to achieve the desired coverage.use_binned_residuals: IfFalse, a single correction factor is applied to all predictions during conformalization. IfTrue, the conformalization process uses a correction factor that depends on the bin where the prediction falls. This can be thought as a type of Adaptive Conformal Predictions and it can lead to more accurate prediction intervals since the correction factor is conditioned on the region of the prediction space.
# Plot intervals
# ==============================================================================
plot_prediction_intervals(
predictions = predictions,
y_true = data_test,
target_variable = "OT",
title = "Predicted intervals",
kwargs_fill_between = {'color': 'gray', 'alpha': 0.4, 'zorder': 1}
)
# Plot intervals with zoom ['2018-05-01', '2018-05-15']
# ==============================================================================
plot_prediction_intervals(
predictions = predictions,
y_true = data_test,
target_variable = "OT",
initial_x_zoom = ['2018-05-01', '2018-05-15'],
title = "Predicted intervals",
kwargs_fill_between = {'color': 'gray', 'alpha': 0.4, 'zorder': 1}
);
# Predicted interval coverage (on test data)
# ==============================================================================
coverage = calculate_coverage(
y_true = data.loc[end_calibration:, 'OT'],
lower_bound = predictions["lower_bound"],
upper_bound = predictions["upper_bound"]
)
print(f"Predicted interval coverage: {round(100 * coverage, 2)} %")
# Area of the interval
# ==============================================================================
area = (predictions["upper_bound"] - predictions["lower_bound"]).sum()
print(f"Area of the interval: {round(area, 2)}")
Predicted interval coverage: 79.32 % Area of the interval: 38409.35
The prediction intervals generated using conformal prediction achieve an empirical coverage very close to the nominal coverage of 80%.
⚠ Warning
Probabilistic forecasting in production
The correct estimation of prediction intervals with conformal methods depends on the residuals being representative of future errors. For this reason, calibration residuals should be used. However, the dynamics of the series and models can change over time, so it is important to monitor and regularly update the residuals. It can be done easily using the set_out_sample_residuals() method.